Recently an article giving an overview of protein coevolution theory is published in the Molecular Biology and Evolution journal by David L. Robertson and Simon C. Lovell from University of Manchester. We can find useful references of current work on this topic such as Hakes(2007), Suel(2003), Socolich(2005), Halabi(2009), Wang and Pollock(2005) and Fares(2006) .
This article talks about the action of evolutionary pressure on the regions of the interacting proteins that contribute to binding. Basically this paper talks about how mutation of a protein at one binding site may lead to mutation of another residue involved in same binding site.
Following figures represents the effect of lowering fitness at one interdependent site leads to increase in fitness of other site.
They also explain the definition of Coevolution from 1950s as "reciprocal evolutionary change in interacting species". There is well stabilized field of evolution(correlation) in RNA structure prediction but in terms of protein structure prediction using coevolution method is still on the stage of development. But it has been proved that coevolving residues are present in many interacting or non-interacting protein domains. We can see an example of the coevolving residues from the same article but is derived from the David Hausslers (2007) article, which calculates coevolving residues based on the the single parametric model of double amino acid pair substitution and then expands their work on whole Pfam database.
An example of sites that demonstrate intermolecular coevolution. (a) Cyanobacterial and (b) human superoxide dismutase. The residues highlighted are at structurally equivalent positions and exhibit strong covariation (Yeang and Haussler 2007). In the proteins shown, the Phe and the Asn/Gln residues have exchanged positions. (c and d) Sequence profile for the equivalent positions. In each case, the cyanobacterialsequence corresponding to panel (a) is at the top and the human sequence corresponding to panel (b) is at the bottom. For other sequences, the Pfam family names are used.
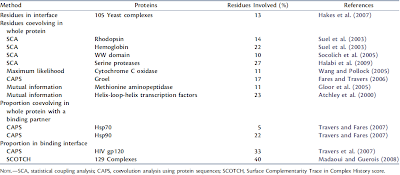
It is known that there are only few residues in each protein domain which may show coevolution. In the next table they have sorted few of the already existing methods for detecting coevolving residues and then compared their prediction of residues involved.
There are several mechanisms that contribute to the degree of correlations of replacements on amino acids either within one protein chain or between chains. Waddell et al. (2007) are explicit: ‘‘correlated evolution is what is detected,
whereas coevolution is the hypothesized cause.’’There are, however, a set of causes that may be hypothesized:
(i) Site-specific coevolution between interacting proteinshas been detected in a range of systems (Moyle et al. 1994; Atchley et al. 2000; Mintseris and Weng 2005; Travers and Fares 2007; Yeang and Haussler 2007; Madaoui and Guerois 2008). It is relatively strong on a ‘‘per-residue’’ basis, indicated by its identification from the analysis of a handful of residues. The signal is most easily detected in the ‘‘rim’’ regions surrounding the interaction interface (Travers and Fares 2007; Yeang and Haussler 2007; Kann et al. 2009) rather than the core of the interface itself (Hakes et al. 2007). This is probably
because the interface itself can be somewhat conserved.
(ii) Correlations of evolutionary rates between interacting proteins when measured over the entire protein length (Williams and Hurst 2000; Fraser et al. 2002). The evidence suggests that these rate correlations are unrelated to coevolution; rather they are due to external factors. This suggestion solves the puzzle of evolutionary correlations between spatial distant sites within protein structures (Hakes et al. 2007) and between proteins that do not directly interact (Juan et al. 2008a). It also explains the relative strength of the observed correlated rates. For obligate complexes (i.e., those that are constitutively bound to their interacting partners), the rate correlation between proteins distant in the complex is as strong as for those directly interacting (Hakes et al. 2007). By contrast, for proteins with a more tenuous functional link, the correlation is much weaker (Juan et al. 2008a).
It is clear that site specific molecular coevolution not only exists but it is also necessary to maintain biological function. Authors argue to improve methods for predicting coevolving residues and they also ask for including the fact that sequences considered for detecting coevolution might have come different origin.


No comments:
Post a Comment